Introduction
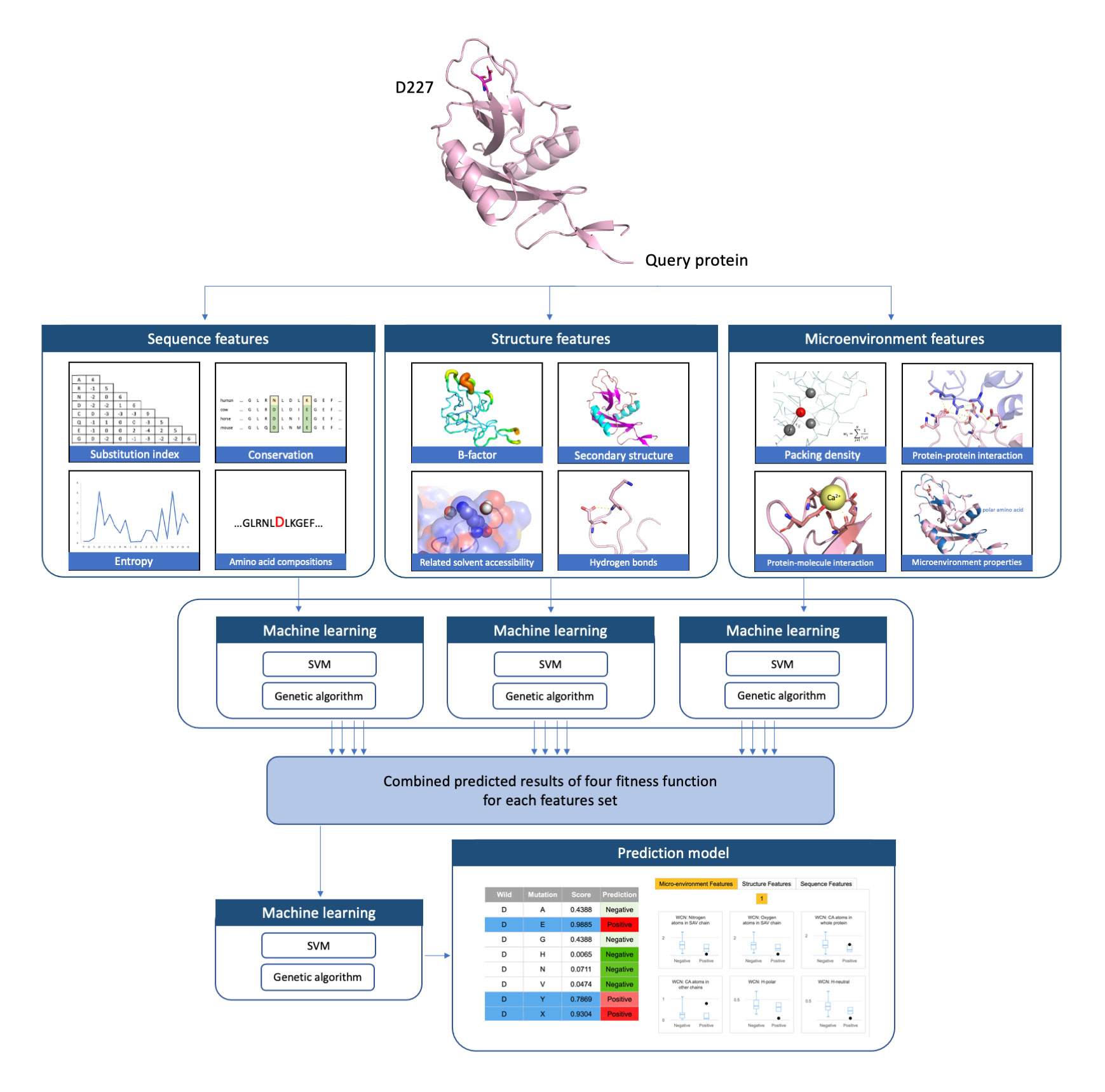
CanSavPre is a structure-based cancer-related single amino acid variation prediction system. This system predicted the cancer-related SAVs and provided the critical features used to estimate the relationship between their properties and cancer caused by SAVs. Moreover, CanSavPre developed by the machine learning methods and its five-fold cross-validation performance is reached 89.73% for accuracy, 0.74 for the Matthews correlation coefficient, and 0.81 for the F1 score.
Why is prediction of cancer-related SAVs important?
Single amino acid variation (SAV) refers to one amino acid substitution resulting from genetic polymorphisms. Increasing evidence indicates that SAVs are associated with several different cancers via structurally or functionally changed. Depend on the mutation position; these SAVs might cause protein destabilization or influence binding affinity and protein-protein interactions resulting in cancer. Our prediction model provides a novel way forward for cancer research, not only for clinical outcomes but also for recognizing prognostic biomarkers, which we contend is a breakthrough for precision medicine.
What is the method of CanSavPre?
We used the machine learning methods to build the cancer-related SAV prediction systems. All the training data were collected from CanProVar2.0. These data were mapped to the protein identified by BLAST from the Protein Data Bank for exacted protein structure of the SAV sequence. Then, the data was split into subgroups to determine the characteristics of specific wild-type amino acid alterations in each of the sequence-based, structure-based and microenvironment-based feature sets. The prediction model used a two-level Support Vector Machine (SVM) classifier module with a genetic algorithm (GA) to select features and optimize performance. Critical descriptors emerge through use of the feature selection procedure. Although further study is needed to reveal the cancer mechanism in most selected features, our results indicate that it is possible to reliably predict cancer-related SAVs.